This is an early preview of PPI.bio. Bugs and missing features are expected. Read more »
 Beta
Beta
How-To Guide for Proteome-Wide PPI Inference
Using the online ppi.bio interface, users are capable of making inferences between:
- a user-defined protein and
- all the proteins of a user-specified organism.
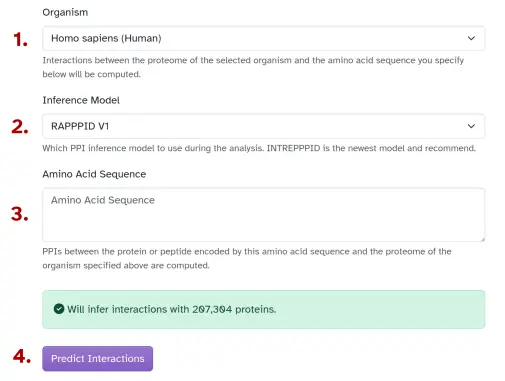
Fill out the "Proteome-Wide Prediction" form
To begin, visit the "Proteome-wide Prediction" page, where you'll be presented with a form asking you for:
- The organism whose proteome to use. Interactions between your submitted amino acid sequence and the proteins of this organism will be computed.
- The inference model to use to infer PPIs. Currently, RAPPPID and INTREPPPID are supported.
- The amino acid sequence of a protein whose interactions you're interesting on inferring. PPIs between the protein encoded by the user-inputted amino acid sequence and the proteome proteins will be inferred.
- Hit "Predict Interactions" to start inferring.
Your task will be added to a queue. Once accepted, a progress bar will appear which shows how your job is going.
Interpreting the Results
Check-out the "Interpreting Results from Proteome-Wide PPI Inference"